Biomolecular Kinetics | Affinity Measurements | Kinetic Screening and Clone Selection | Epitope Binning | Isotyping | Select Binding Pairs
Biomolecular Kinetics
Kinetic analysis of antibodies and other proteins is critical to both clone selection and characterization in development and production. The Octet® family of instruments accurately measures kinetic constants by bringing the detection surface directly to the sample, eliminating the need for micro-fluidics. This unique approach utilizing label-free, real-time analysis streamlines laboratory workflow and expedites assay development. It allows direct measurement of crude samples while minimizing instrument maintenance.
Key Features of Kinetics on the Octet Systems
- Kinetic analysis: easily and accurately determine ka, kd, and KD
- Kinetic screening: 96 or 384 measurements with full kinetic profiles, not just end-point
- Increased throughput: up to 96 simultaneous reads
- Assay directly in crude samples: no need for sample purification or preparation
Affinity Measurements — ka, kd, KD
Unlike rough estimates of kinetic information from IC50 values obtained via ELISAs, real-time kinetic measurements offer a direct and more realistic depiction of molecular interactions. Each instrument in the Octet family is a highly capable label-free system for full affinity determination, enabling users to easily and accurately obtain kinetic constants such as ka, kd, KD. The measurement below illustrates a full kinetic characterization using streptavidin biosensors. A titration series of the antigen is measured against an immobilized antibody. The data set is analyzed with global fitting, thereby producing a ka, kd, KD.
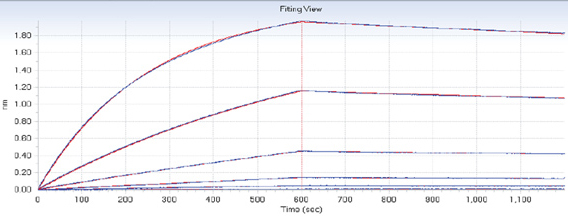
Figure 1. Kinetic characterization of an antibody-antigen interaction, blue curves represent experimental data and red curves represent the statistical fitting of curves.
Kinetic Screening and Clone Selection
Researchers looking to select the best clones from a large pool will reap substantial benefits with the Octet family of instruments due to their throughput and crude sample compatibility. As we move into a new era of drug discovery, the number of samples or clones to be tested will continue to increase and effectively screening them will become more crucial.
Affinity Screening
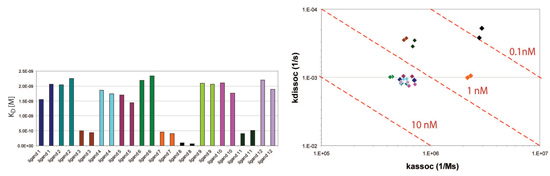
In an affinity screen of receptor-ligand interactions, lysate targets were assayed against a series of amine-coupled receptors using the Octet system. A purified receptor was immobilized onto the amine reactive sensor and assayed against twelve ligands in duplicate. The receptor-ligand screen in Figure 2 illustrates good reproducibility between replicate kinetic values for all twelve ligands. An affinity isotherm plot of the association and dissociation constants is used to further discriminate interactions that have equal affinity (KD) but have different kinetic properties.
Figure 2: Kinetic characterization of the receptor:ligand interaction. The affinity (KD) bar chart and kd/ka isotherm plot is presented. Click image for larger view.
Off-rate Screening
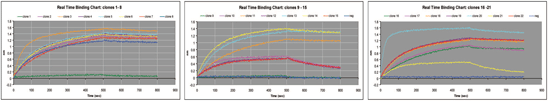
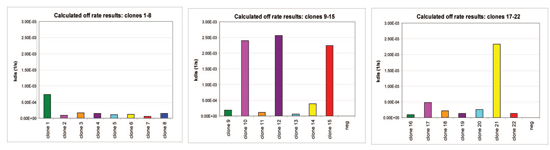
Using Streptavidin biosensors on the Octet platform, a biotinylated antigen was immobilized onto the biosensor surfaces offline. Twenty-two clones were screened against the antigen for binding and subsequent off-rate analysis. The antigen was immobilized for 500 seconds, and a 300-second off-rate was subsequently assayed in a single run. Figure 3 shows the actual real-time kinetic binding charts for three sets of eight sensors sampled (N=22). The Octet system software calculates and graphs the off-rates (Kd); enabling rapid identification of the clones' rank order.
Figure 3A: Real-time binding charts of three series of 8 biosensors, comprising a screen of 22 clones to the biotinylated antigen. Click image for larger view.
Figure 3B: Matched off-rate charts of the screen of 22 clones to the biotinylated antigen. Click image for larger view.
Epitope Binning
When multiple candidates have similar affinities for a target, determining their preference for the same or different epitopes on the target molecule becomes a crucial factor in their evaluation. Separating the candidates into groups (bins) with a common preference for the same epitope helps researchers further characterize the specificity of the clones and their ability to block target activity, which can have far reaching effects on the candidate's efficacy and pharmacokinetics.
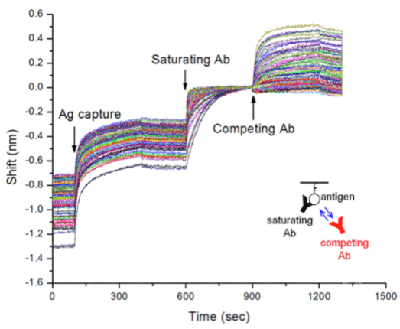
Figure 4: Binning of 192 antibodies in 5 hours. Abdiche et. al. Rinat-Pfizer. In a high-throughput binning experiment, the antigen is captured onto the biosensor followed by binding the first antibodies to saturation. Then binding of a competing antibody is measured against the bound pair. Positive binding signal from the competing antibody indicates lack of competition, i.e. the first antibody and the competing antibody bind to different epitopes on the antigen.
Isotyping
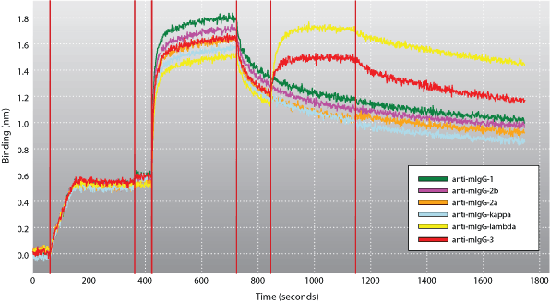
Detecting different antibody isotypes in the presence of background proteins is quick and easy using the Octet system. A biosensor immobilized with an anti-species antibody will capture that particular antibody from a crude mixture, such as a hybridoma supernatant. Exposure to a series of isotype-specific antibodies provides a sandwich format for quickly identifying the antibody's isotype out of a crude mixture.
Figure 5: A biotinylated anti-mouse Fc antibody was immobilized onto Streptavidin biosensors. The biosensors were then used to bind the mIgG (10 μg/mL) to be isotyped out of a sample with a high-protein background (> 1mg/mL). After a brief wash in buffer, the biosensors were exposed to a series of isotype-specific antibodies as noted in the legend. By taking advantage of the Octet system's ability to process up to eight samples simultaneously, a panel of isotype-specific antibodies was used to detect the mIgG's isotype in ~15 minutes. Click figure for larger image.
Select Binding Pairs
Screening for potential binding pairs using an Octet system takes a fraction of the time it would take using traditional methods. Optimal binding pair selection can greatly reduce the time needed to create new binding assays such as ELISAs. By immobilizing a single ligand onto eight biosensors, the binding of any analytes loaded into a 96-well sample plate can quickly be detected. Once the assay is complete, a qualitative assessment of the results will identify which analytes bound best to the immobilized ligand.
Figure 6 illustrates the real-time binding chart of the antibody pair screen. Two pairs can be immediately identified as suitable for further assay development. Ab1 capture antibody with detection antibody Ab5 exhibits good binding to the analyte. The interaction between Ab1/Ab5 also exhibits some dissociation, indicating a weak affinity. Ab2 capture antibody with Ab4 detection antibody exhibits both good binding and strong affinity.
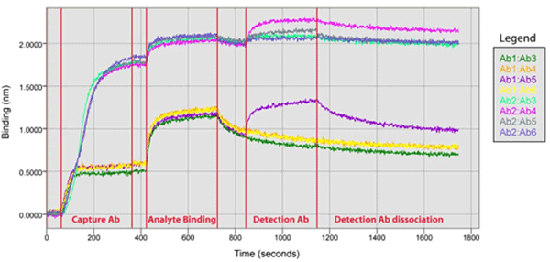
Figure 6: